Variational Autoencoding for Biologists
- 2 minsInspired by Greg Johnson’s Integrated Cell paper on generative modeling of cellular structure, I spent a couple days exploring variational autoencoders to derive useful latent spaces in biological data. I’ve found that I often learn best when preparing to teach. To that aim, I wrote a tutorial on VAEs in the form of a Colab notebook working through mathematical motivations and implementing a simple model. The tutorial goes on to play with this model on some of the Allen Institute for Cell Science data.
VAEs may seem a bit far afield from biological data analysis at first blush. Without repeating too much of the tutorial here, I find a few specific properties of these models particularly interesting:
1 - VAE latent spaces are continuous and allow for linear interpolation.
This means that operations like $z_\text{Metaphase} = z_\text{Prophase} + z_\text{Anaphase}/2$ followed by decoding to the measurement space often yield sane outputs.
Biological Use Case: Predict unseen intermediary data in a timecourse experiment.
2 - VAEs are generative models, providing a lens from which we can begin to disentangle individual generative factors beneath observed variables.
High dimensional biological data, like transcriptomes or images, is the emergent product of underlying generative factors. Think of a generative factor as a semantically meaningful parameter of the process that generated your data. In the case of cell biology, these factors may be aspects of cellular state, environmental variables like media conditions, or time in a dynamic process. Recent work from DeepMind et. al. on $\beta$-VAEs has shown promise for finding latent dimensions that map to specific generative factors, allowing for interpretable latent dimensions.
Biological Use Case: Learn a latent space where generative factors of interest like cell cycle state, differentiation state, &c., map uniquely to individual dimensions, allowing for estimates of covariance between generative factors and measurement space variables.
3 - VAE latent spaces provide a notion of variation.
When encoding a given observation, VAEs provide not only a location in the latent space, but an estimate of variance around this mean. This estimate of variation provides a metric of mapping confidence that’s not only useful for estimating the likelihood of alternative outcomes, but can be used for more general tasks like anomaly detection.
Biological Use Case: Run an image based screen and use a VAE model trained on control samples to estimate which perturbations deviate from the control distribution.
Take a spin through the tutorial if you’re so inclined. As a teaser, you get to generate interesting figures like this:
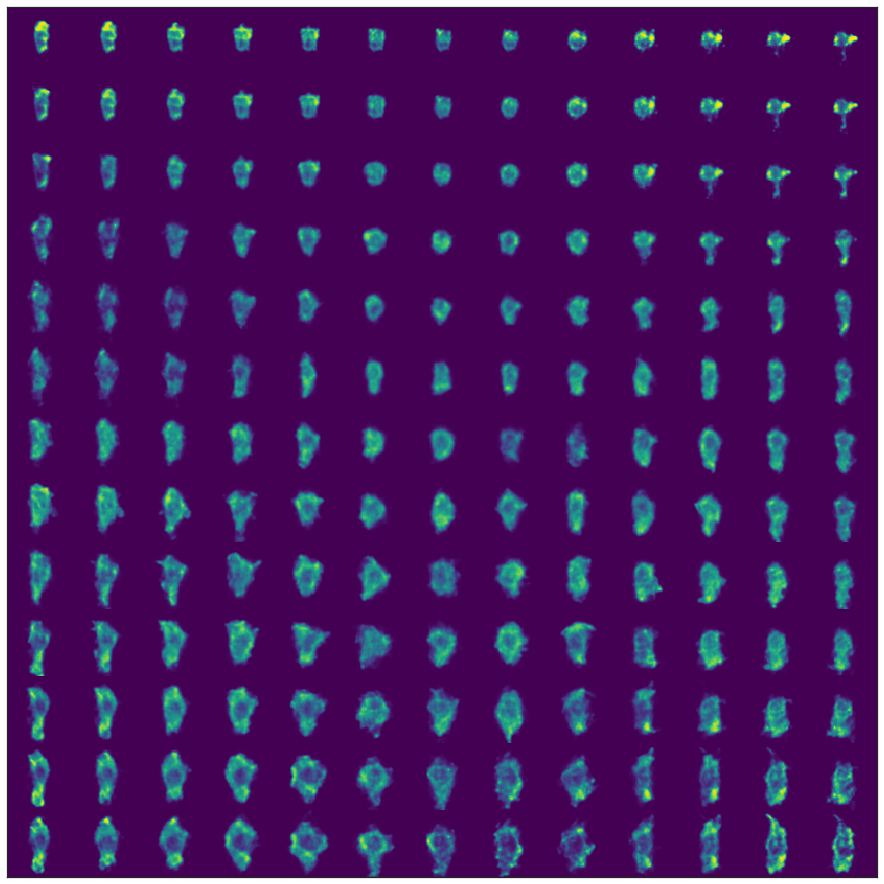
If you see any issues or hiccups, please feel free to email me.
